Welcome to OncoSplicing!
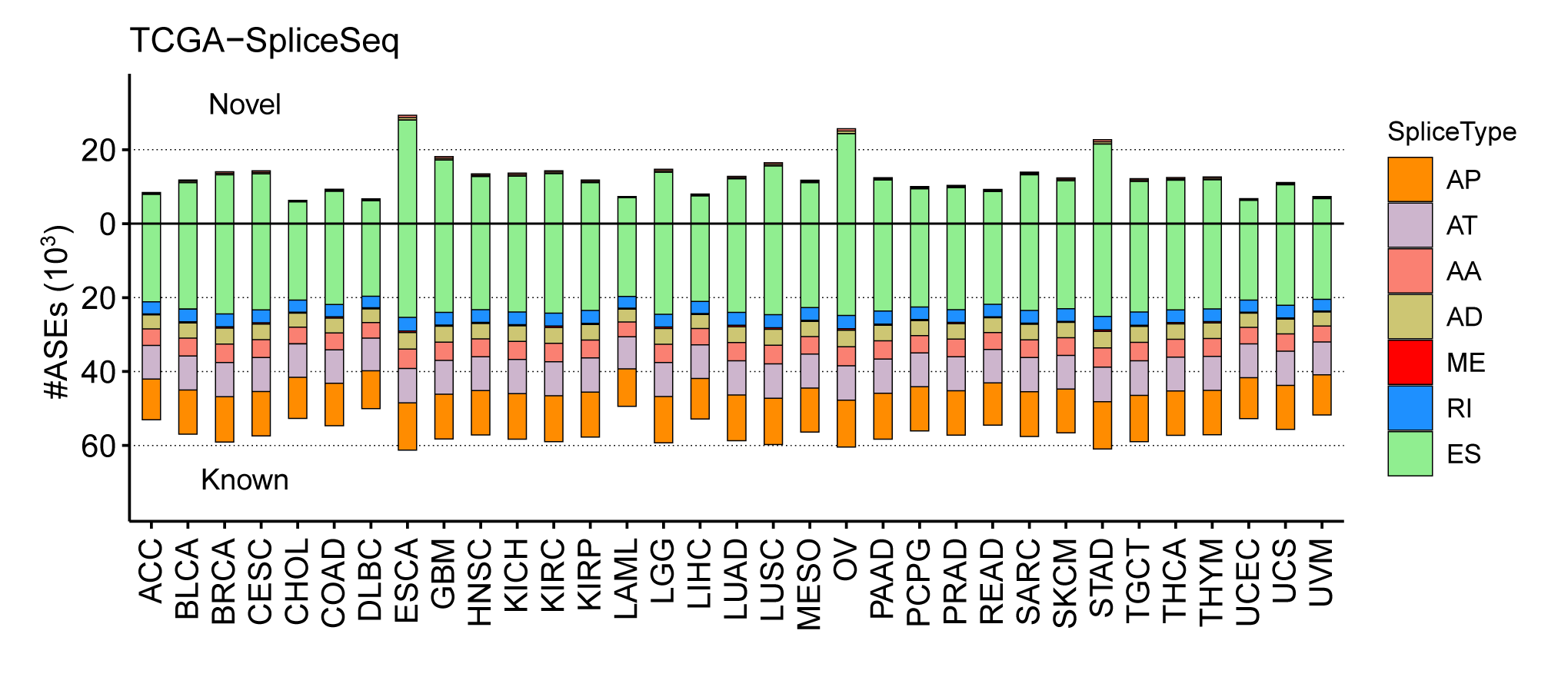
OncoSplicing is a database to systematically study clinically relevant alternative splicing in 33 TCGA cancers and 31 GTEx tissues. Alternative splicing (AS) represents a crucial method in mRNA level to regulate gene expression, and contributes to the mRNA processing and protein complexity. Many researches have indicated that alternative splicing could be potential biomarkers for cancer diagnosis and drug targets for treatment. We developed the first version of OncoSplicing in 2019 to view survival associated and differential AS events in cancers, based on a majority of splicing data from the TCGA SpliceSeq database. Recently, we added splicing data of 31 GTEx tissues as well as 32 TCGA cancers from the SplAdder project, which contained much more novel AS events. In summary, a total of 122,423 AS events in the SpliceSeq project and 238,558 AS events in the SplAdder project were included in this database.
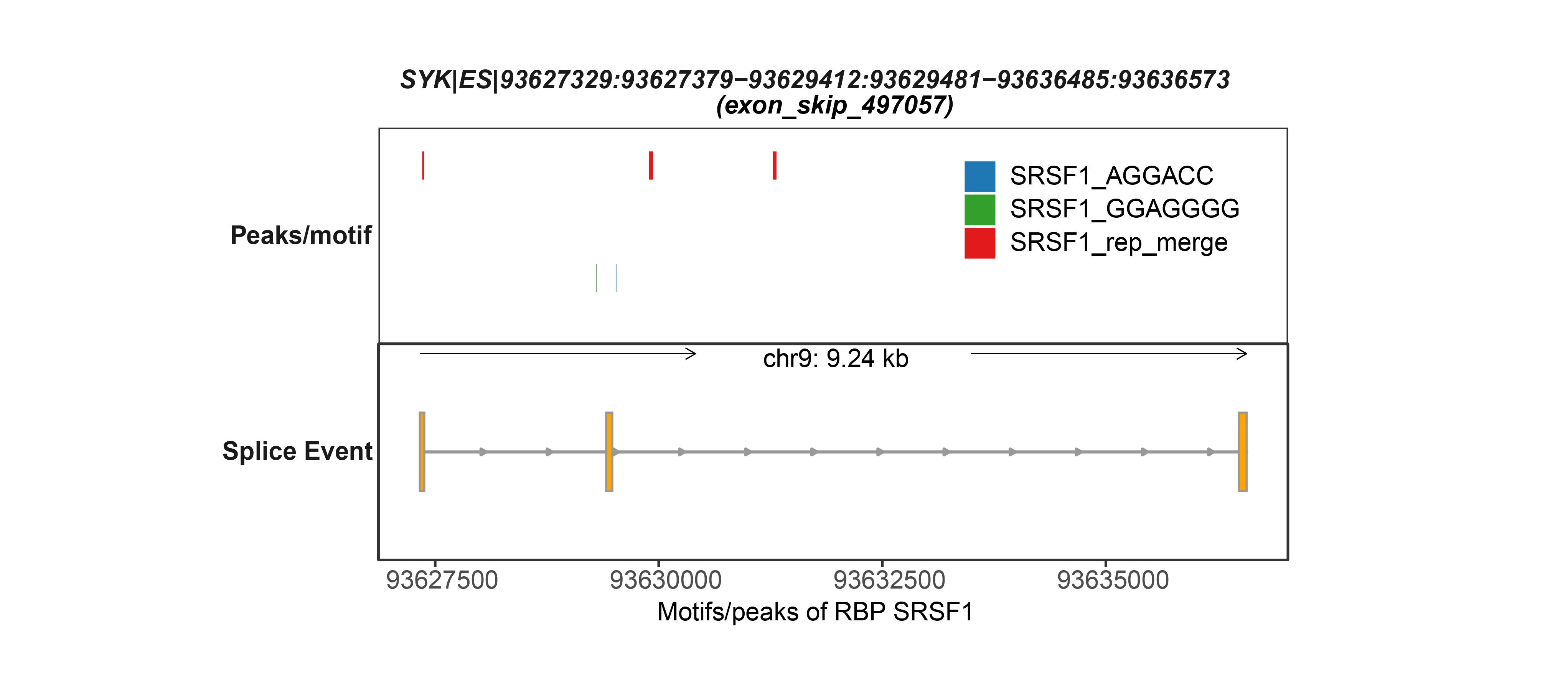
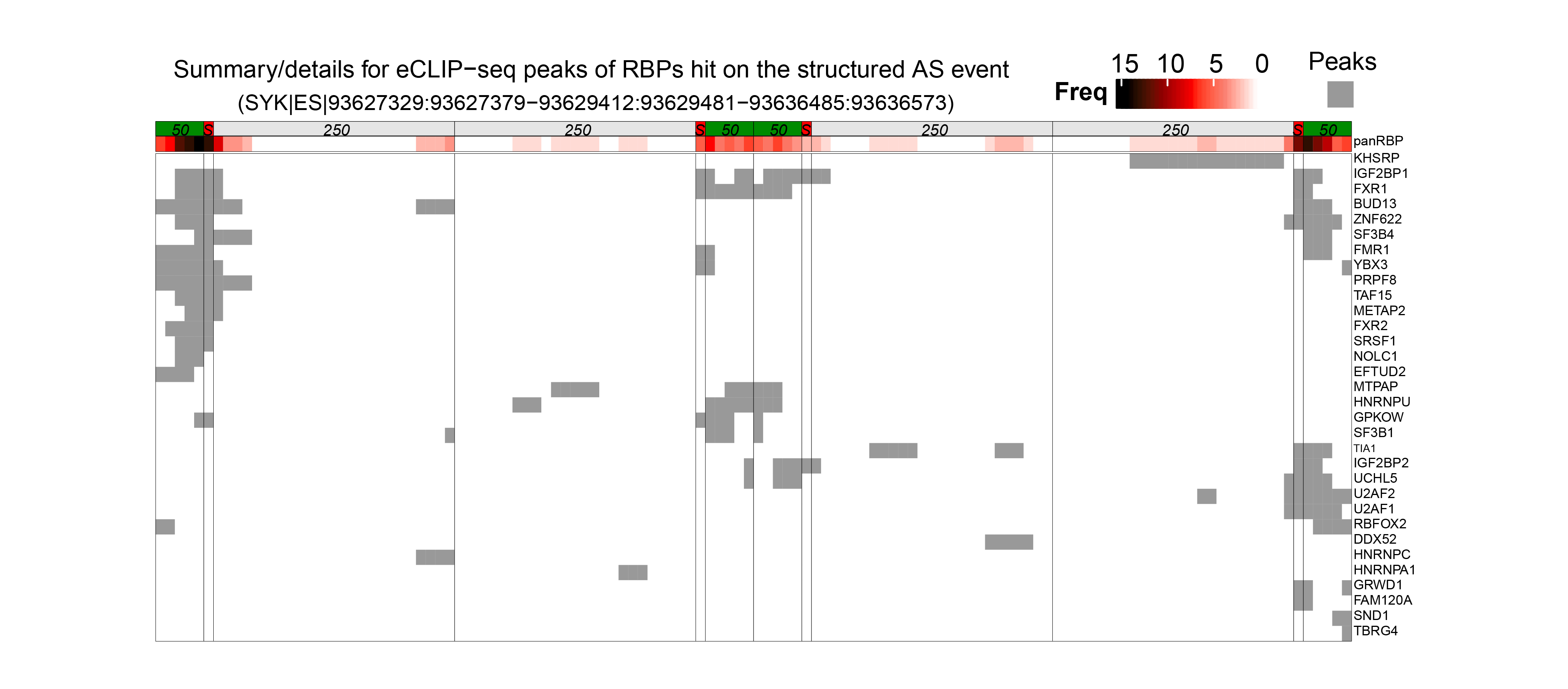
In the latest version of OncoSplicing, we integrated 282,125 splice events from two sources: 57,172 SpliceSeq events involving a single alternative exon, and 238,558 SplAdder events. We then mapped these events with RNA-binding motifs for 130 RBPs, eCLIP peaks for 150 RBPs, and splice events regulated by 358 RBPs from the ENCODE project. Correlation analyses were conducted for all integrated events with 753 unique RBPs, including both the collected RBPs and an additional 521 splicing regulators annotated in MsigDB. As a result, we identified 58,993,935 (27.8% of all) AS event-RBP pairs with at least one regulatory relationship, and 1,412,583 (0.7% of all) were classified as high-confidence.
In OncoSplicing, users can:
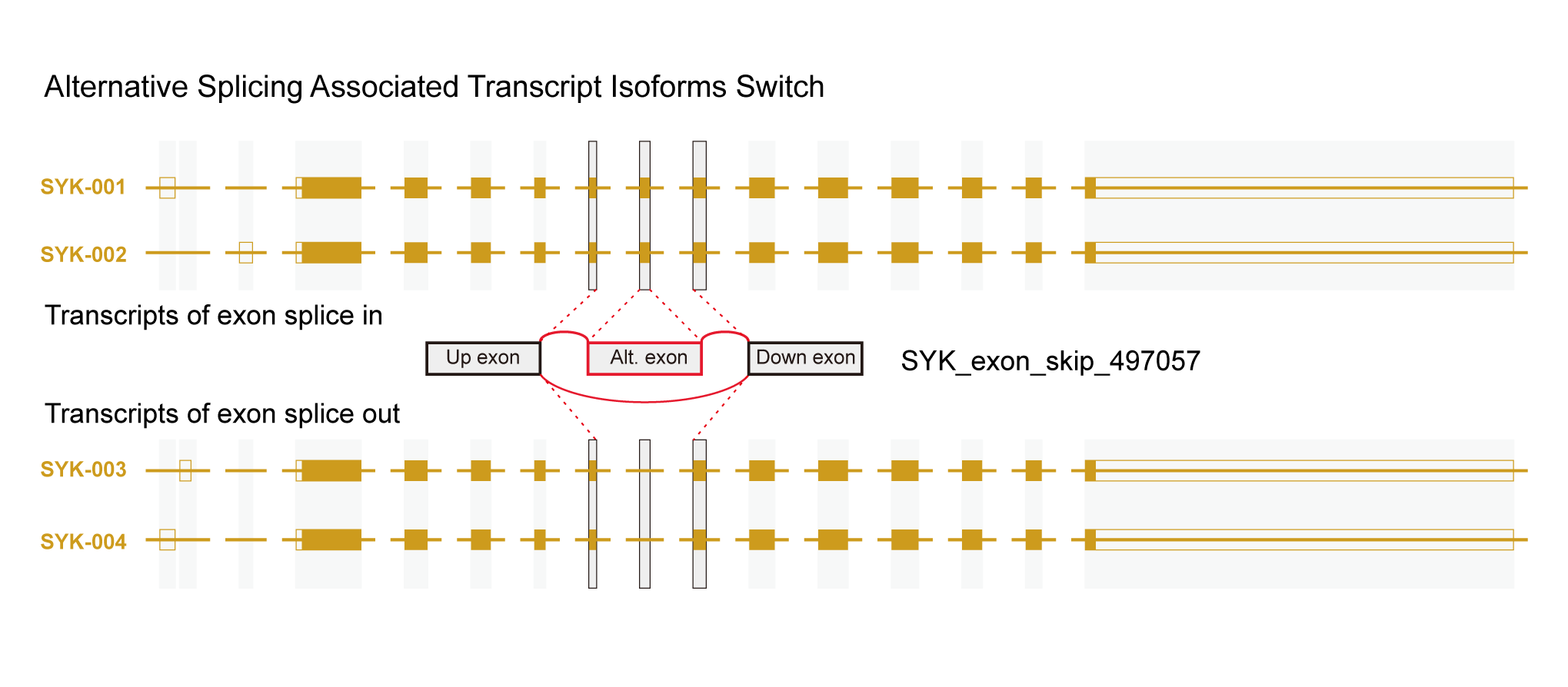
1. Browse or search a gene symbol for integrative AS information of the queried gene, including chromosomal location, AS associated transcripts and statistic results;
2. Browse or search a gene symbol in a cancer type to identify survival associated and/or differential AS events and to perform visualization;
3. Browse or search a gene symbol and select a splice event for pan-cancer views of PSI distribution in TCGA cancers and/or GTEx tissues and results of survival and/or differential analyses;
4. Browse or search a gene symbol in a cancer type to identify clinical indicator relevant AS events;
5. Download all results of analysis, figures of querying and PSI data in the SplAdder project for further integrative study.
In the latest OncoSplicing, users can also:
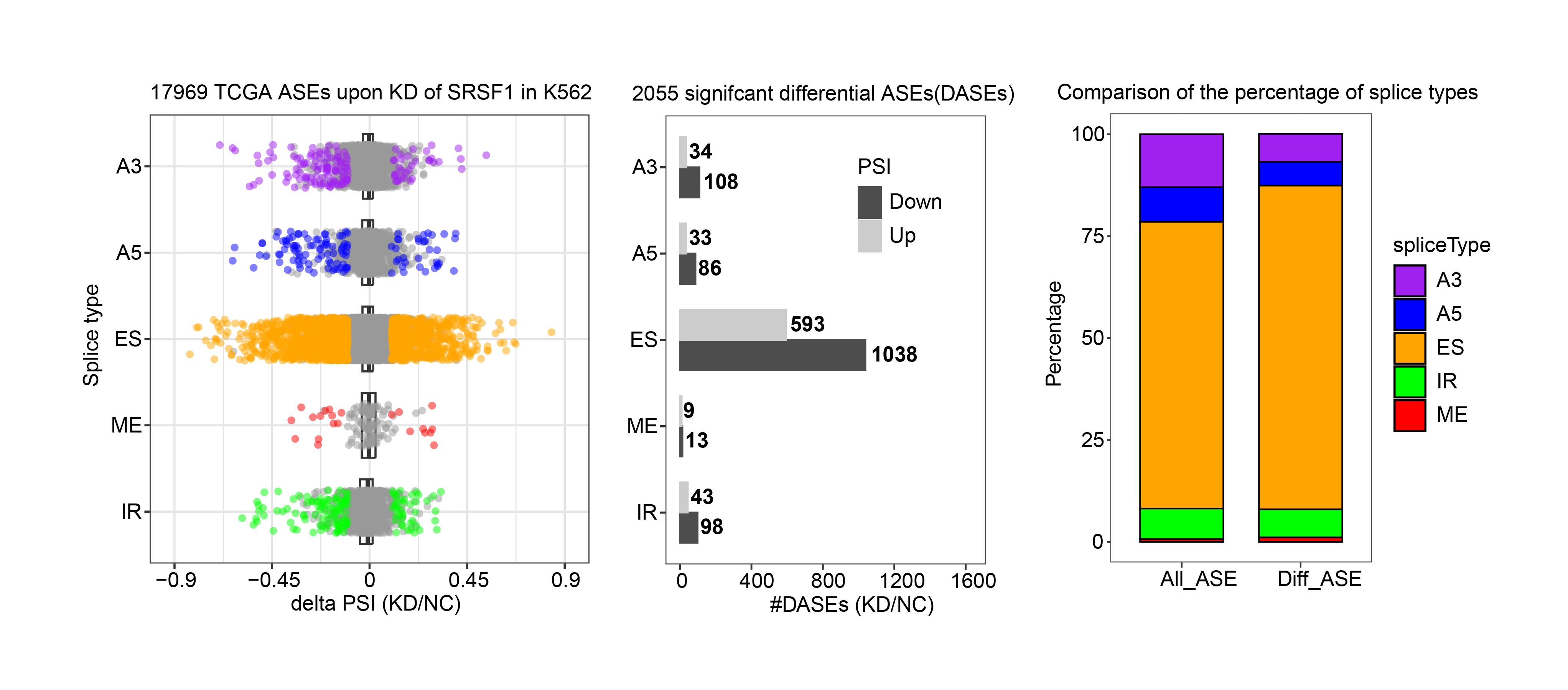
6. Search by gene symbol, AS event, or RBP to explore integrative splicing relationships between AS events and hundreds of RBPs.
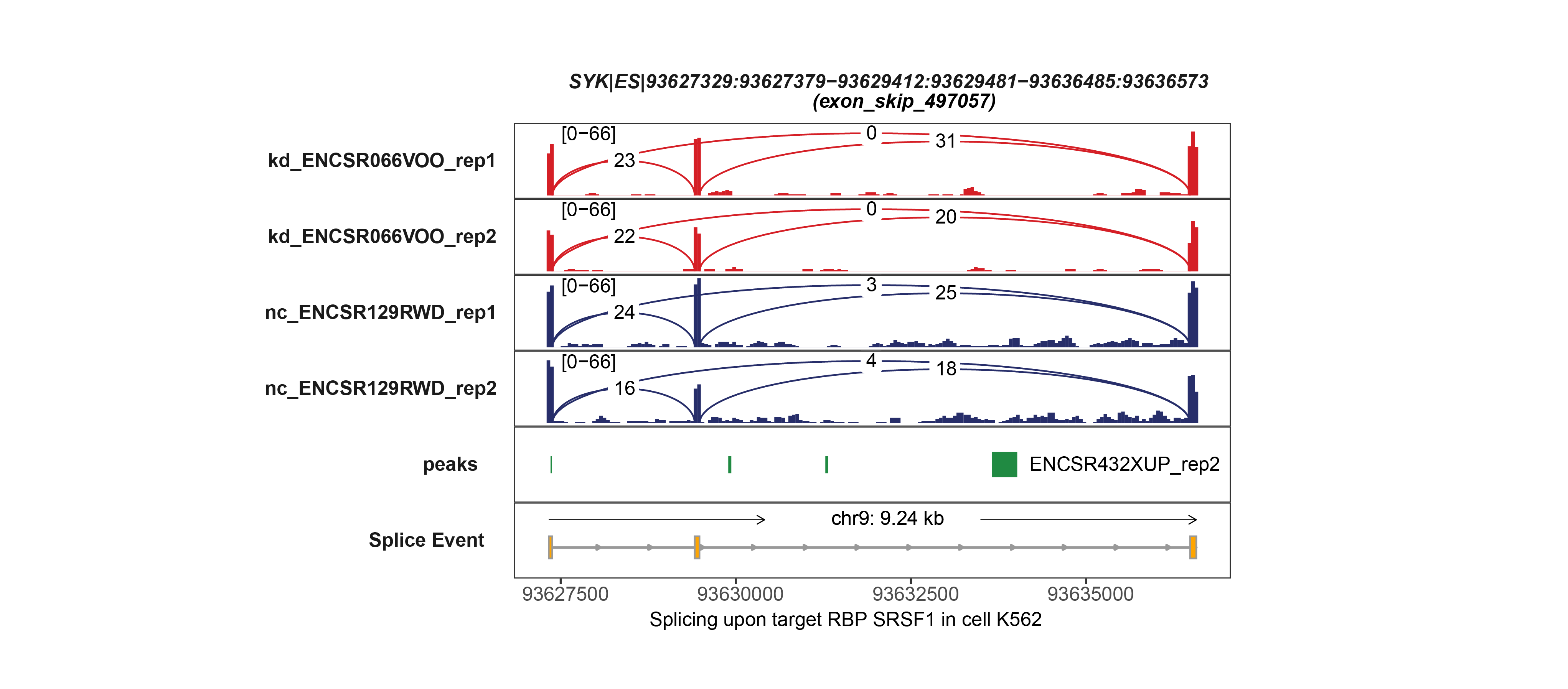
7. Search by gene symbol, AS event, or RBP to investigate regulatory relationships between AS events and hundreds of RBPs from the ENCODE project.
8. Conduct co-expression analysis between different omics data, including splicing data and mRNA expression of RBPs, across TCGA and/or GTEx cohorts.